Transformers For Molecular Generation
🌈 Abstract
The article discusses the use of transformer networks for generating molecular structures based on desired physical properties. It covers the key components of the transformer network, including the multi-head attention mechanism, handling of varying sequence lengths, and the overall architecture. The article also presents an implementation of the transformer model in PyTorch and evaluates the model's performance on generating SMILES strings for molecules.
🙋 Q&A
[01] The Transformer Network; Multihead-Attention
1. What is the key component at the heart of the transformer network?
- The key component at the heart of the transformer network is the multi-head attention mechanism, which enables the model to focus on all parts of the sequence at the same time.
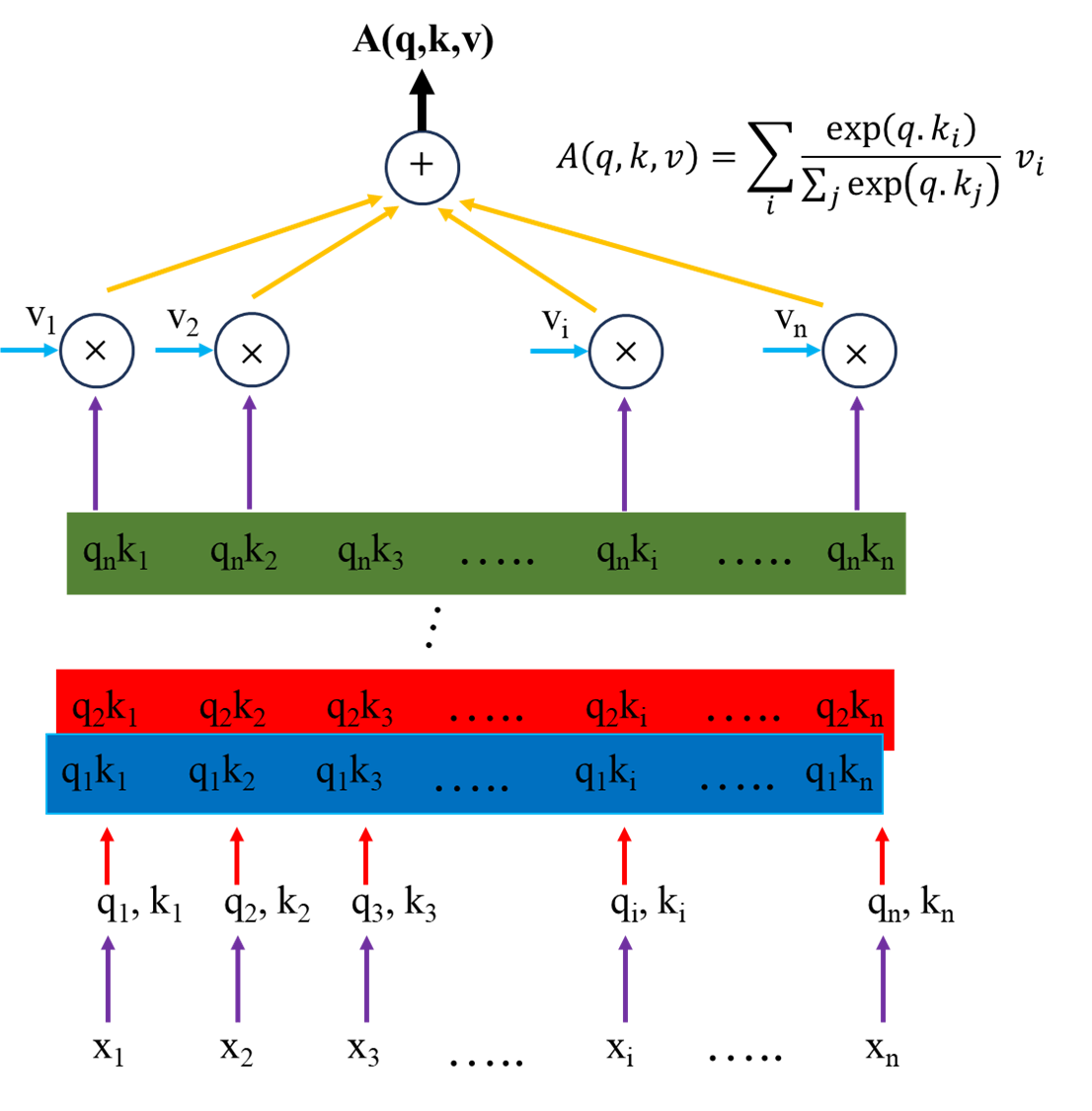
2. How does the multi-head attention mechanism work?
- The multi-head attention mechanism is built upon three main components derived from the input sequence: query (q), key (k), and value (v). The dot-product attention (q.k) is used to compute the attention scores between each query and all keys, and these attention scores are passed through a softmax function to obtain the attention weights. The attention weights and the values matrix are then used to determine an attention layer (also referred to as a head).
3. What is the purpose of using multiple heads in the attention mechanism?
- Using multi-head attention allows the model to run the attention mechanism described above multiple times, each with different learnable matrices (i.e., different Wq, Wk, Wv). This enables the model to focus on different aspects of the input sequence simultaneously.
[02] Masks: Managing Varying Sequence Lengths
1. What is the purpose of the padding mask?
- The padding mask helps the network pay attention only to the actual molecule and not the padding, by assigning a large negative attention score to the padding tokens.
2. What is the purpose of the look-ahead mask?
- The look-ahead mask is used during training to prevent the model from accessing future tokens in a sequence, ensuring that predictions of each token in the sequence are made based only on past and present tokens.
[03] The Whole Architecture
1. How does the encoder layer of the transformer model differ from the traditional transformer encoder?
- Since the molecular properties input is not a sequence, the traditional encoder of the transformer network is replaced with a simple ANN feed-forward network.
2. How does the decoder layer of the transformer model work?
- The decoder layer follows the same architecture as the traditional transformer decoder, with two multi-head attention layers, a feed-forward network, and layer normalization in-between.
3. What are the two key elements used in the decoder?
-
- The target sequence is converted to an embedding using random embedding vectors.
-
- A positional encoding is used to include the sequential information of the sequence, as the multi-head attention network does not inherently know the relative or absolute positions of tokens.
[04] Evaluating The Model
1. What are some of the strengths and limitations of the generated molecules?
- The model is able to generate chemically valid SMILES strings, demonstrating that it has learned some underlying principles of molecular structure. However, it struggles with capturing specific stereochemical configurations and tends to generate simpler molecules compared to the target structures.
- It is unclear how well the generated molecules match the desired input molecular properties, as the evaluation focuses more on structural similarity rather than property matching.
2. What are some potential reasons for the model's limitations?
- The limited training data or the need to adjust the model architecture to better capture stereochemical information and generate more complex molecular structures.
- The challenge of mapping the input molecular properties to unique molecular structures, which may require additional feature engineering or a different modeling approach.
</output_format>